A story about #ExpressScan
Glad this preprint by stellar 💫 MD-PhD Joost Mekke on the #ExpressScan in the Athero-Express from the strategic Circulatory Health theme is finally out.
It's been a long and winding road. Here's the story👇🏽.
First a little background
Cardiovascular diseases CVD are the leading cause of death in the world; check out this great graphic 👇🏽 - that sums it up nicely. Atherosclerosis is the primary underlying mechanism of CVD leading to plaque formation in the arteries.
Cardiovascular disease
WHO statistics show cardiovascular disease is the world’s number 1 killer.
Athero-Express
Since 2002 we have collected over 3,600 plaques in the Athero-Express. We paraffin-embed, cut, and stain these plaques for commonly present cells and structures ('phenotypes'), and manually study plaques through microscopy 🔬
In 2010 we found that intraplaque hemorrhage (IPH) associates with secondary outcome after carotid endarterectomy (CEA)👇🏽.
In 2013 we found that [sex/gender] associates with IPH and modulates the association with secondary outcome after CEA, more in men ♂ than women ♀👇🏽. This work was led by Joyce Vrijenhoek and Hester den Ruijter and marked the start of research into sex differences in CVD at the UMC Utrecht.
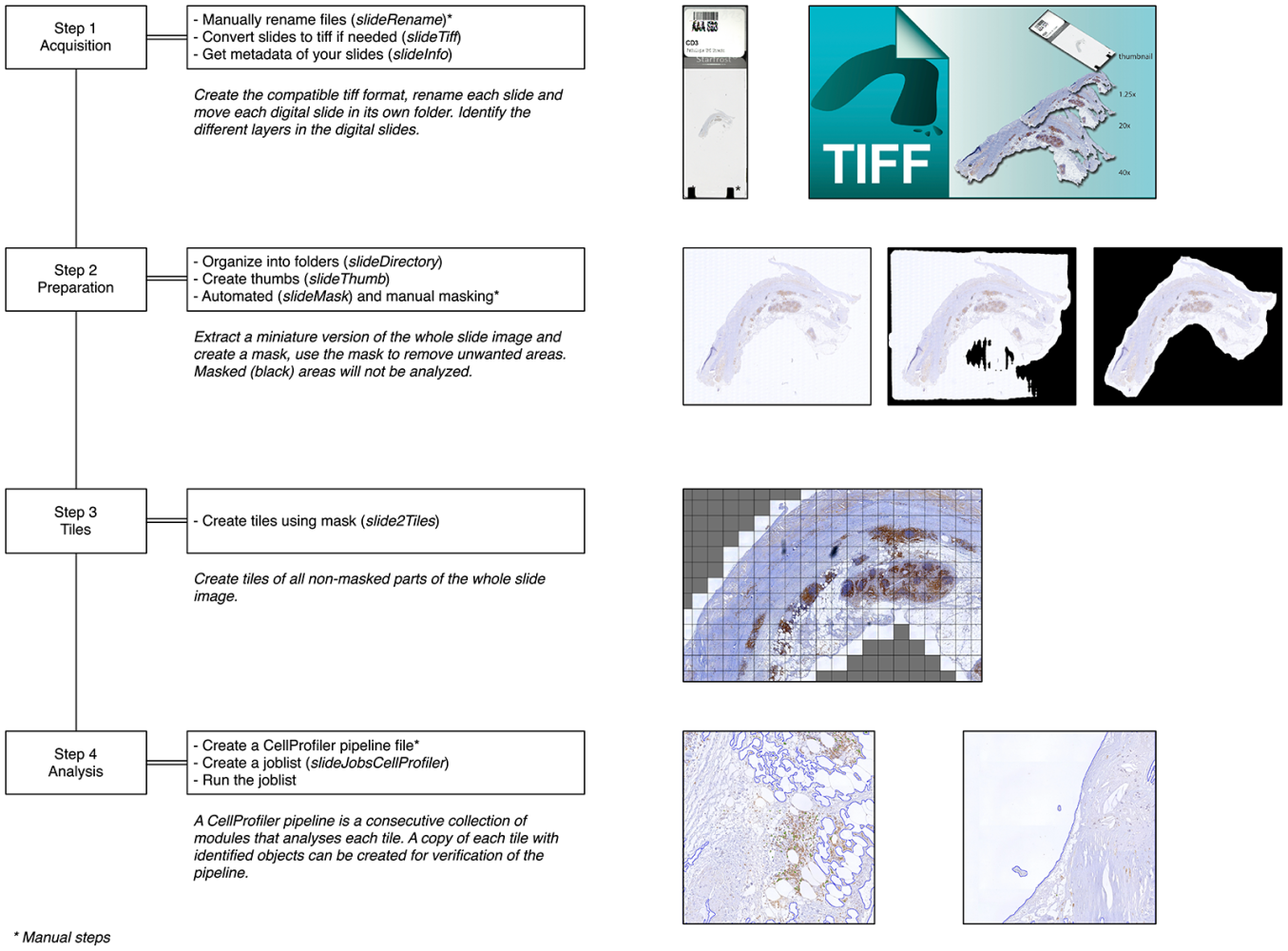
#ExpressScan
But manual analysis is cumbersome 😣😓😮💨. So back in 2014 💫 MD/coder Bas Nelissen created slideToolKit: a collection of scripts that enable fast, reproducible, and automated processing of high-throughput whole-slide images (WSI) for analysis with CellProfiler. You can find the GitHub here and be sure to read the original paper in PLoS One.
slideToolKit: a collection of scripts to enable fast, reproducible, and automated processing of whole-slide images.
Since then we have been crawling through all the slide cabinets 🤪, cleaned the slides🧼🧽, and used #pathology scanners to obtain over 25,000 high-resolution WSI for all plaques. This totals up to about 20Tb 😬. A hell of a job involving multiple people, microscopic lamps breaking down, and pathology scanners from Roche and Hamamatsu.
"Oh, but Sander, what does it look like 🧐?", you might wonder. Here's an example, this sample was stained (the brown colouring) for CD68 a protein that is expressed by macrophages (left), and ACTA2 which is expressed by smooth muscle cells (right).
Glycophorin C
Fast forward to 2021. Glycophorin C (GLYCC) is a glycoprotein found on the membrane of red blood cells and a presumed marker of the degree of IPH. We quantified GLYCC using slideToolKit and CellProfiler, and investigated the relation of GLYCC and six other markers with symptoms and secondary major adverse cardiovascular events (MACE).
Here are the 3️⃣ main findings 👇🏽
1️⃣ The total area of positive GLYCC stain was larger in individuals with manually scored IPH, and more so in men ♂ than women ♀.
2️⃣ As IPH is one of the causes of plaque rupture, we assessed the correlation of GLYCC with symptoms prior to CEA (upper graph, below).
3️⃣ GLYCC associates with secondary MACE after CEA (lower graph, below).
The future is bright
But, for me this 7 year story does not end with the current preprint on GLYCC in plaques. Here's why 👇🏽
☝🏽Automated, high-throughput quantification of WSI is easy with slideToolKit; it takes less than a day to analyse all WSI 👊🏽.
✌🏽WSI quantification opens up a whole range of new analyses 🧐🔬🧑🏽🔬👨🏻🔬👩🏼🔬🥼🧫.
Some ideas💡we are working on:
💡 genotype-phenotype analyses 👉🏽 another 💫 PhD Kai Cui is working on this, stay tuned for her presentation at the ESHG 2021 😀.
💡 discovery of clinically relevant features in WSI through machine learning 👉🏽 the ✨PhD Yipei Song is working with me, Clint Miller, and Craig Glastonbury and to finish CONVOCALS and DEEPENIGMA.
💡 In the spirit of Open Science #ExpressScan should be Open Access. That's easier said than done. First step is to open it up to the wider CirculatHealth community together with Lennart Landsmeer. So watch this space for more.
Finally
It's been a long time coming, with a lot of hurdles 🙈🙉🙊to overcome, but I am very happy with this first milestone of #ExpressScan 🤩🥳. The preprint is open for massive open peer review 👉🏽 https://bit.ly/ExpressScan and the codes 💻are 👉🏽https://bit.ly/ExpressScanCode.
A big thank you 🤗 to all the co-authors (in no particular order) Clint Miller, Gerard Pasterkamp, Hester den Ruijter, Michal Mokry, Tim Sakkers, Yipei Song, Noortje van den Dungen, Dominique de Kleijn, Maarten Verwer, Gert Jan de Borst. A special thank you 🙇🏽should go to Saskia Haitjema and Joost Mekke for pulling this off and co-leading this part of the project! And of course we owe a debt of gratitude 🙏🏽 to all those that worked on #ExpressScan in the past: Bas Nelissen (#ExpressScan would not have been possible without his work), Joyce Vrijenhoek, Marijke Linschoten, Tim van de Kerkhof, Robin Reijers, Tim Bezemer, Sander Reukema, Joelle van Bennekom, and many more. Lastly, we should acknowledge and be grateful to the hundreds of patients willing to participate in the Athero-Express, without their support and altruism there is no scientific progress.